Comparison to SOTA¶
This notebook contains a comparison of the Vietoris-Rips (VR) persistent homology (PH) computation (on the GPU) implemented in torchph to the implementations available via dionysus (v2) and ripser (which is SOTA in terms of runtime).
This runtime study is a small-scale version of the runtime study in
Connectivity-Optimized Representation Learning via Persistent Homology
C. Hofer, R. Kwitt, M. Dixit and M. Niethammer
ICML ’19
[1]:
import sys
[2]:
# LOCALLY INSTALL RIPSER
!{sys.executable} -m pip install ripser
Requirement already satisfied: ripser in /scratch4/chofer/opt/anaconda/envs/pyt_1.4/lib/python3.7/site-packages (0.4.1)
Requirement already satisfied: numpy in /scratch4/chofer/opt/anaconda/envs/pyt_1.4/lib/python3.7/site-packages (from ripser) (1.18.1)
Requirement already satisfied: scipy in /scratch4/chofer/opt/anaconda/envs/pyt_1.4/lib/python3.7/site-packages (from ripser) (1.4.1)
Requirement already satisfied: persim in /scratch4/chofer/opt/anaconda/envs/pyt_1.4/lib/python3.7/site-packages (from ripser) (0.1.2)
Requirement already satisfied: Cython in /scratch4/chofer/opt/anaconda/envs/pyt_1.4/lib/python3.7/site-packages (from ripser) (0.29.15)
Requirement already satisfied: scikit-learn in /scratch4/chofer/opt/anaconda/envs/pyt_1.4/lib/python3.7/site-packages (from ripser) (0.22.1)
Requirement already satisfied: matplotlib in /scratch4/chofer/opt/anaconda/envs/pyt_1.4/lib/python3.7/site-packages (from persim->ripser) (3.1.3)
Requirement already satisfied: hopcroftkarp in /scratch4/chofer/opt/anaconda/envs/pyt_1.4/lib/python3.7/site-packages (from persim->ripser) (1.2.5)
Requirement already satisfied: joblib>=0.11 in /scratch4/chofer/opt/anaconda/envs/pyt_1.4/lib/python3.7/site-packages (from scikit-learn->ripser) (0.14.1)
Requirement already satisfied: pyparsing!=2.0.4,!=2.1.2,!=2.1.6,>=2.0.1 in /scratch4/chofer/opt/anaconda/envs/pyt_1.4/lib/python3.7/site-packages (from matplotlib->persim->ripser) (2.4.6)
Requirement already satisfied: python-dateutil>=2.1 in /scratch4/chofer/opt/anaconda/envs/pyt_1.4/lib/python3.7/site-packages (from matplotlib->persim->ripser) (2.8.1)
Requirement already satisfied: kiwisolver>=1.0.1 in /scratch4/chofer/opt/anaconda/envs/pyt_1.4/lib/python3.7/site-packages (from matplotlib->persim->ripser) (1.1.0)
Requirement already satisfied: cycler>=0.10 in /scratch4/chofer/opt/anaconda/envs/pyt_1.4/lib/python3.7/site-packages (from matplotlib->persim->ripser) (0.10.0)
Requirement already satisfied: six>=1.5 in /scratch4/chofer/opt/anaconda/envs/pyt_1.4/lib/python3.7/site-packages (from python-dateutil>=2.1->matplotlib->persim->ripser) (1.14.0)
Requirement already satisfied: setuptools in /scratch4/chofer/opt/anaconda/envs/pyt_1.4/lib/python3.7/site-packages (from kiwisolver>=1.0.1->matplotlib->persim->ripser) (45.2.0.post20200210)
[3]:
# LOCALLY INSTALL DIONYSUS
!{sys.executable} -m pip install dionysus
Requirement already satisfied: dionysus in /scratch4/chofer/opt/anaconda/envs/pyt_1.4/lib/python3.7/site-packages (2.0.6)
[4]:
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
from collections import defaultdict
# IMPORT RIPSER and DIONYSUS
from ripser import ripser
import dionysus as d
import torch
import time
from scipy.spatial.distance import pdist
from sklearn import datasets
from torchph.pershom import vr_persistence_l1, vr_persistence
device = 'cuda'
[5]:
def gen_random_10D_data(n):
return np.random.randn(n,10)
def gen_circlces(n):
return datasets.make_circles(
n_samples=n,
noise=0.05,
factor=0.5)[0]
N = np.arange(50,500,50)
times = defaultdict(list)
f = gen_random_10D_data
for n in N:
trials = defaultdict(list)
print('Evaluating N={}'.format(n))
for i in np.arange(50):
#Generate data
x = f(n)
D_l1 = pdist(x, metric='cityblock')
thr_l1 = np.max(D_l1.ravel())
X = torch.Tensor(x).to(device)
# Timing torchph
t0 = time.time()
l, _ = vr_persistence_l1(X.contiguous(),0, 0);
t1 = time.time()
trials['torchph'].append(t1-t0)
if 'ripser' in sys.modules:
t0 = time.time()
ripser(x, maxdim=0, thresh=thr_l1, metric='manhattan')
t1 = time.time()
trials['ripser'].append(t1-t0)
if 'dionysus' in sys.modules:
t0 = time.time()
dd = pdist(x, metric='cityblock')
filt = d.fill_rips(dd, 1, thr_l1)
m = d.cohomology_persistence(filt)
t1 = time.time()
trials['dionysus'].append(t1-t0)
times[n] = trials
Evaluating N=50
Evaluating N=100
Evaluating N=150
Evaluating N=200
Evaluating N=250
Evaluating N=300
Evaluating N=350
Evaluating N=400
Evaluating N=450
[6]:
methods = []
if 'ripser' in sys.modules:
methods.append('ripser')
if 'dionysus' in sys.modules:
methods.append('dionysus')
methods.append('torchph')
T = np.zeros((len(N),3))
S = np.zeros((len(N),3))
for j,(k,v) in enumerate(times.items()):
for i, method in enumerate(methods):
T[j,i] = np.array(v[method]).mean()
S[j,i] = np.array(v[method]).std()
[7]:
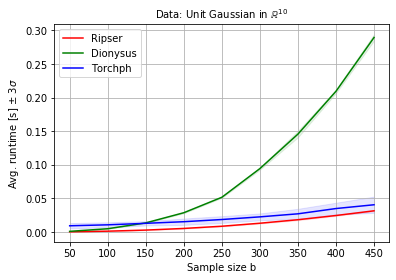
colors = ['red', 'green', 'blue']
plt.figure(figsize=(6,4))
for i, method in enumerate(methods):
plt.plot(N, T[:,i], label=method.title(), color=colors[i])
plt.fill_between(N, T[:,i]-3*S[:,i], T[:,i]+3*S[:,i], color=colors[i], alpha=0.1)
plt.legend()
plt.grid()
plt.xlabel('Sample size b')
plt.ylabel(r'Avg. runtime [s] $\pm$ 3$\sigma$')
plt.title(r'Data: Unit Gaussian in $\mathbb{R}^{10}$', fontsize=10)
plt.savefig('/tmp/normal10d_runtime.pdf', bbox_inches='tight')
Run a simple sanity check that all VR PH computations return the same result.
[8]:
# Sample data
x = f(200)
# Compute l1 distance matrix and get max. pairwise distance = threshold
D_l1 = pdist(x, metric='cityblock')
thr_l1 = np.max(D_l1.ravel())
# Run Ripser
dgm_ripser = ripser(x, maxdim=0, thresh=thr_l1, metric='manhattan')['dgms']
# Run Ours
X = torch.Tensor(x).to(device)
l, _ = vr_persistence_l1(X.contiguous(),0, 0);
# Run Dionysus
filt = d.fill_rips(D_l1, 1, thr_l1)
m = d.cohomology_persistence(filt)
dgms = d.init_diagrams(m, filt)
[9]:
assert(np.abs(dgm_ripser[0][:,1][:-1] - l[0].cpu().numpy()[:,1]).sum() < 1e-4)
assert(np.abs(sorted([x.death for x in dgms[0]])[:-1] - l[0].cpu().numpy()[:,1]).sum() < 1e-4)
assert(np.abs(dgm_ripser[0][:,1][:-1] - sorted([x.death for x in dgms[0]])[:-1]).sum() < 1e-4)